Module 1.0 - Mini-ML¶
Model¶
Models: parameterized functions.
- $m(x; \theta)$
- $x \text{ - input}$
- $m \text{ - model}$
Initial Focus:
- $\theta \text{ - parameters}$
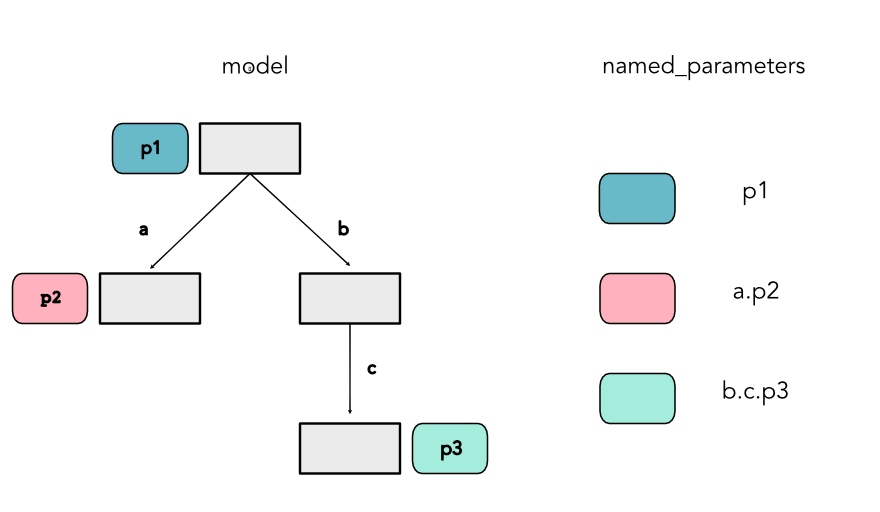
Specifying Parameters¶
- Datastructures to specify parameters
- Requirements
- Independent of implementation
- Compositional
Module Example¶
In [2]:
from minitorch import Module, Parameter
class OtherModule(Module):
pass
class MyModule(Module):
def __init__(self):
# Must initialize the super class!
super().__init__()
# Type 1, a parameter.
self.parameter1 = Parameter(15)
# Type 2, user data
self.data = 25
# Type 3. another Module
self.sub_module = OtherModule()
Module Naming¶
Lecture Quiz¶
Quiz
Outline¶
Model
Parameters
Loss
Datasets¶
Data Points¶
- Convention $x$
In [3]:
split_graph([s1[0]], [])
Out[3]:
Data Points¶
- Convention $x$
In [4]:
split_graph([s1[1]], [])
Out[4]:
Data Points¶
- Convention $x$
In [5]:
split_graph(s1, [])
Out[5]:
Data Labels¶
- Convention $y$
In [6]:
split_graph([s1[0]], [s2[0]])
Out[6]:
Training Data¶
- Set of datapoints, each $(x,y)$
In [7]:
split_graph(s1, s2)
Out[7]:
Data Set¶

Model¶
Models¶
- Functions from data points to labels
- Functions $m(x; \theta)$
- Any function is okay (e.g. Modules)
Example Model¶
Example of a simple model
x = (0.5, 0.2)
In [8]:
@dataclass
class Model:
def forward(self, x):
return 0 if x[0] < 0.5 else 1
Model 1¶
- Linear Model
In [9]:
from minitorch import Parameter, Module
class Linear(Module):
def __init__(self, w1, w2, b):
super().__init__()
self.w1 = Parameter(w1)
self.w2 = Parameter(w2)
self.b = Parameter(b)
def forward(self, x1: float, x2: float) -> float:
return self.w1.value * x1 + self.w2.value * x2 + self.b.value
Model 1¶
In [10]:
model = Linear(w1=1, w2=1, b=-0.9)
draw_graph(model)
Out[10]:
Model 2¶
In [11]:
class Split:
def __init__(self, linear1, linear2):
super().__init__()
# Submodules
self.m1 = linear1
self.m2 = linear2
def forward(self, x1, x2):
return self.m1.forward(x1, x2) * self.m2.forward(x1, x2)
In [12]:
model_b = Split(Linear(1, 1, -1.5), Linear(1, 1, -0.5))
draw_graph(model_b)
Out[12]:
Model 3¶
In [13]:
class Part:
def forward(self, x1, x2):
return 1 if (0.0 <= x1 < 0.5 and 0.0 <= x2 < 0.6) else 0
In [14]:
draw_graph(Part())
Out[14]:
Parameters¶
Parameters¶
- Knobs that control the model
- Any information that controls the model shape
Parameters¶
- Change $\theta$
In [15]:
show(Linear(w1=1, w2=1, b=-0.5))
show(Linear(w1=1, w2=1, b=-1))
Out[15]:
Linear Parameters¶
a. rotating the linear separator
In [16]:
model1 = Linear(w1=1, w2=1, b=-1.0)
model2 = Linear(w1=0.5, w2=1.5, b=-1.0)
In [17]:
compare(model1, model2)
Out[17]:
Linear Parameters¶
b. changing the separator cutoff
In [18]:
model1 = Linear(w1=1, w2=1, b=-1.0)
model2 = Linear(w1=1, w2=1, b=-1.5)
In [19]:
compare(model1, model2)
Out[19]:
In [20]:
def forward(self, x1: float, x2: float) -> float:
return self.w1 * x1 + self.w2 * x2 + self.b
Loss¶
What is a good model?¶
In [21]:
hcat([show(Linear(1, 1, -1.0)), show(Linear(1, 1, -0.5))], 0.3)
Out[21]:
Distance¶
- $|m(x)|$ correct or incorrect
In [22]:
with_points(s1, s2, Linear(1, 1, -0.4))
Out[22]:
Points¶
In [23]:
pts = [
with_points([s1[0]], [], Linear(1, 1, -1.5)),
with_points([s1[0]], [], Linear(1, 1, -1)),
with_points([s1[0]], [], Linear(1, 1, -0.5)),
]
hcat(pts, 0.3)
Out[23]:
Loss¶
- Loss weights our incorrect points
- Uses distance from boundary
$L(w_1, w_2, b)$ is loss, function of parameters.
Warmup: ReLU¶
In [24]:
def point_loss(m_x):
return minitorch.operators.relu(m_x)
In [25]:
graph(point_loss, [], [])
Out[25]:
Loss of points¶
In [26]:
graph(point_loss, [], [-2, -0.2, 1])
Out[26]:
Loss of points¶
In [27]:
graph(lambda x: point_loss(-x), [-1, 0.4, 1.3], [])
Out[27]:
Full Loss¶
In [28]:
def full_loss(m):
l = 0
for x, y in zip(s.X, s.y):
l += point_loss(-y * m.forward(*x))
return -l
Point Loss¶
In [30]:
graph(point_loss, [], [])
Out[30]:
In [31]:
graph(point_loss, [], [-2, -0.2, 1])
Out[31]:
What is a good model?¶
In [32]:
hcat([show(Linear(1, 1, -1.0)), show(Linear(1, 1, -0.5))], 0.3)
Out[32]:
Sigmoid Function¶
In [33]:
graph(minitorch.operators.sigmoid, width=8).scale_x(0.5)
Out[33]: